-Search query
-Search result
Showing all 31 items for (author: rossmann & fm)

EMDB-11059: 
ATP-dependent partner switch links flagellar C-ring assembly with gene expression
Method: subtomogram averaging / : Blagotinsek V, Schwan M, Steinchen W, Mrusek D, Hook J, Rossmann FM, Freibert SA, Kressler D, Beeby M, Thormann KM, Bange G

EMDB-11060: 
Flagellar motor of Shewanella putrefaciens in situ, flhG deletion
Method: subtomogram averaging / : Blagotinsek V, Schwan M, Steinchen W, Mrusek D, Hook J, Rossmann FM, Freibert SA, Kressler D, Beeby M, Thormann KM, Bange G

EMDB-10943: 
In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog
Method: subtomogram averaging / : Rossmann FM, Hug I, Sangermani M, Jenal U, Beeby M

EMDB-10945: 
In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog
Method: subtomogram averaging / : Rossmann FM, Hug I, Sangermani M, Jenal U, Beeby M
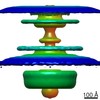
EMDB-10949: 
In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog
Method: subtomogram averaging / : Rossmann FM, Hug I, Sangermani M, Jenal U, Beeby M

EMDB-10950: 
In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog
Method: subtomogram averaging / : Rossmann FM, Hug I, Sangermani M, Jenal U, Beeby M
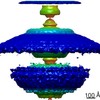
EMDB-10955: 
In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog
Method: subtomogram averaging / : Rossmann FM, Hug I, Sangermani M, Jenal U, Beeby M

EMDB-10956: 
In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog
Method: subtomogram averaging / : Rossmann FM, Hug I, Sangermani M, Jenal U, Beeby M
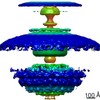
EMDB-10957: 
In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog
Method: subtomogram averaging / : Rossmann FM, Hug I, Sangermani M, Jenal U, Beeby M

EMDB-10057: 
Plesiomonas shigelloides polar motor
Method: subtomogram averaging / : Ferreira JF, Beeby M

EMDB-4569: 
Flagellar motor relic structure from Plesiomonas shigelloides
Method: subtomogram averaging / : Ferreira JF, Beeby M

PDB-3j0b: 
cryo-EM reconstruction of West Nile virus
Method: single particle / : Zhang W, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG

EMDB-5296: 
cryo-EM reconstruction of West Nile virus
Method: single particle / : Zhang W, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG

EMDB-5445: 
Icosahedral reconstruction of the Streptococcus phage C1 capsid
Method: single particle / : Aksyuk AA, Bowman VD, Kaufmann B, Fields C, Klose T, Holdaway HA, Fischetti VA, Rossmann MG

EMDB-5446: 
Asymmetric reconstruction of the Streptococcus phage C1
Method: single particle / : Aksyuk AA, Bowman VD, Kaufmann B, Fields C, Klose T, Holdaway HA, Fischetti VA, Rossmann MG
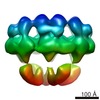
EMDB-2056: 
Electron cryo-microscopy of the Hantaan virus glycoprotein spike
Method: single particle / : Battisti AJ, Chu YK, Chipman PR, Kaufmann B, Jonsson CB, Rossmann MG

PDB-3iyw: 
West Nile virus in complex with Fab fragments of MAb CR4354 (fitted coordinates of envelope proteins and Fab fragments of one icosahedral ASU)
Method: single particle / : Rossmann MG, Kaufmann B

EMDB-5190: 
West Nile Virus in complex with Fab fragments of the neutralizing monoclonal antibody CR4354
Method: single particle / : Kaufmann B, Vogt MR, Holdaway HA, Goudsmit J, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG

EMDB-1624: 
Three-dimensional structure of the recombinant, virus-like particle of Penaeus stylirostris densovirus (PstDNV) or infectious hypodermal and hematopoietic necrosis virus (IHHNV)
Method: single particle / : Kaufmann B, Bowman VD, Li Y, Szelei J, Tijssen P, Rossmann MG

EMDB-5115: 
West Nile virus in complex with a single-chain antibody derivative of the neutralizing monoclonal antibody E16
Method: single particle / : Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Kuhn RJ, Diamond MS, Rossmann MG

PDB-3ixx: 
The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53
Method: single particle / : Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH

PDB-3ixy: 
The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53
Method: single particle / : Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH

EMDB-5102: 
Immature Dengue Virus (DENV) in complex with Fab fragments of the anti-fusion loop antibody E53
Method: single particle / : Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH

EMDB-5103: 
Immature West Nile Virus (WNV) in complex with Fab fragments of the anti-fusion loop antibody E53
Method: single particle / : Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH

EMDB-1466: 
Human Parvovirus B19 (DNA-containing wildtype)
Method: single particle / : Kaufmann B, Chipman PR, Modrow S, Rossmann MG

EMDB-1467: 
Human Parvovirus B19 (empty wildtype particle)
Method: single particle / : Kaufmann B, Chipman PR, Modrow S, Rossmann MG

EMDB-1468: 
Virus-like particle of Human Parvovirus B19 (VLP consisting of VP2 only)
Method: single particle / : Kaufmann B, Chipman PR, Modrow S, Rossmann MG

EMDB-1326: 
Minute virus of mice, a parvovirus, in complex with the Fab fragment of a neutralizing monoclonal antibody.
Method: single particle / : Kaufmann B, Lopez-Bueno A, Garcia-Mateu M, Chipman PR, Nelson CDS, Parrish CR, Almendral JM, Rossmann MG

EMDB-1314: 
Structure of immature West Nile virus.
Method: single particle / : Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG

PDB-2of6: 
Structure of immature West Nile virus
Method: single particle / : Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG

EMDB-1234: 
West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody.
Method: single particle / : Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
